Resources
Phi-Pipe
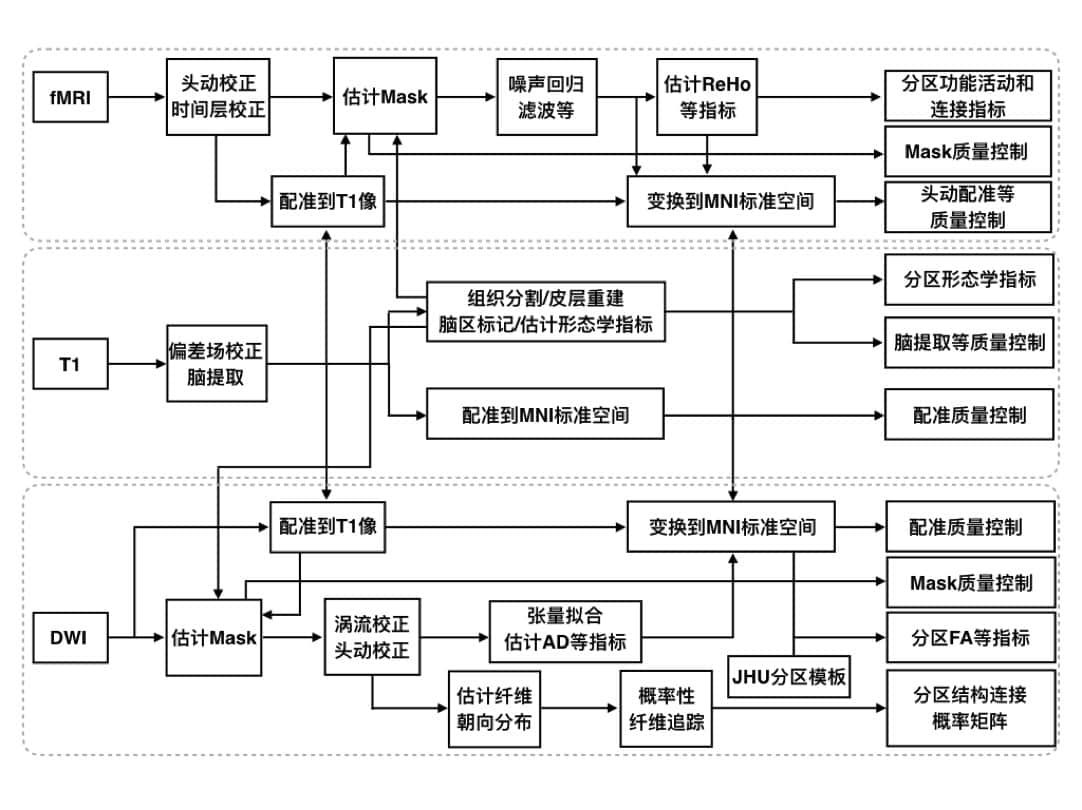
The Phi-Pipe is a multi-modal brain MRI data processing pipeline developed in PHI. Remarkable features of this pipeline include:
- A multi-modal pipeline for preprocessing of T1-3D, resting-state fMRI, and DWI data. This is a one-stop solution if you have scanned multi-modal images. Supported modality combinations are T1, T1+rest, and T1+DWI.
- Supports automatic parallel computing. Make good use of computational power to automatically accelerate the processing.
- Gives multiple levels of results. You can get preprocessed images for your customized further analyses or just tables of parcel-wise measures (such as parcel-wise cortical thickness, reho, and functional connectivity matrices) for your statistical comparisons without touching the images.
- Makes quality control and quality check easy. The pipeline automatically evaluates and reports the quality of your data, and it generates figures for your visual check.
- Supports web-interface. We have implemented an easy-to-use website in our center, called WeProMRI, where users can do “one-click image preprocessing”

The source code of the basic version of PHI-pipe can be downloaded at github release page Here is a user’s manual.
Reference paper Details of the PhiPipe as well various validations and comparisons are documented in our paper.
Reliability and Validity Database We further evaluated the test-retest reliability and predictive validity of PhiPipe’s brain features. We provide an online database to make the access to the reliability and validity measures easier. Users can query the reliability and validity measures for specific brain regions and image features here: PhiPipe Reliability and Validity Database
Brain Growth Charts for Preschool Children
The preschool brain growth chart provides a standardized model for the individualized and refined assessment of the golden period of brain development, and is a basic tool for children’s brain research. A “brain development score” inferred from brain growth charts also helps in early identification of a variety of brain developmental disorders.
Our research team is dedicated to constructing children’s brain growth curves to implement individualized brain examinations and “brain development score” assessments. The team constructed a growth curve model of a total of 90 brain morphological features to assess the relative position of each brain structure in children within the same age and same sex population, revealing the developmental abnormalities of certain brain structures. We will continue to collect data and update these growth charts.
We share the following resources:
- The latest growth charts: Preschool Brain Growth Charts;
- R objects of the growth charts, for automated inferrence of “brain development score” for each feature;
- Sex-stratified brain templates for each year between the ages of 1-6.
gRAICAR
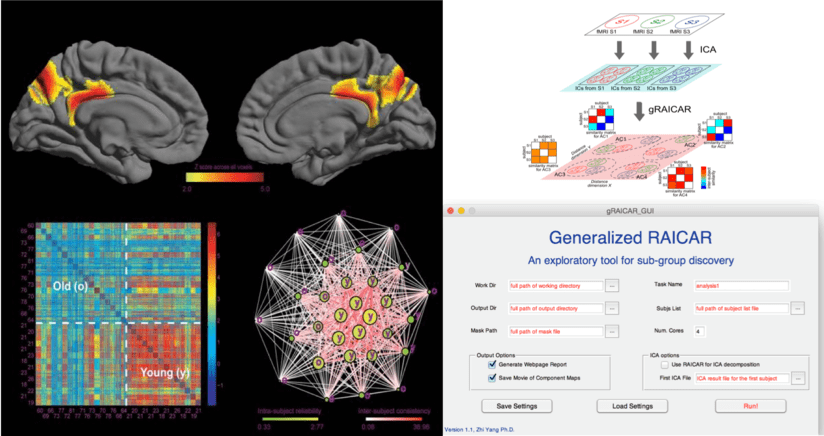
gRAICAR is a Matlab package for data-driven subject community detection based on functional networks.
When we cannot assume that all subjects share the same functional networks, we may want to look into the inter-subject similarity on each network. gRAICAR provides a tool for realizing such a purpose. gRAICAR first decomposes fMRI data into spatial components using independent component analysis (ICA), then aligns the spatial maps across the subjects, yielding an inter-subject similarity matrix for each aligned, group-level component. According to many ICA studies, some of these aligned components represent functional connectivity networks.
The gRAICAR inter-subject similarity matrices reveal potential differences among subjects and thus accelerate data-driven discovery. Further, community detection algorithms applied to the gRAICAR intersubject similarity matrices can provide more quantitative metrics to characterize the associations between individual differences in functional networks and behaviors.
The gRAICAR code and a tutorial with examples: Github link
Please consider to cite the following publications:
Original algorithms:
- Yang Z, Zuo X, Wang P, Li Z, Laconte S, Bandettini PA, Hu X (2012). Generalized RAICAR: Discover homogeneous subject (sub)groups by reproducibility of their intrinsic connectivity networks. NeuroImage 63, 403-414 full text
- Yang Z, LaConte S, Weng X, and Hu X (2008). Ranking and averaging independent component analysis by reproducibility (RAICAR). Human Brain Mapping 29, 711-25 full text
Improvements and applications:
- Yang Z, Chang C, Xu T, Jiang L, Handwerker D, Castellanos F, Milham M, Bandettini P, Zuo X (2014). Connectivity Trajectory across Lifespan Differentiates the Precuneus from the Default Network. NeuroImage 89, 45-56. full text
- Yang Z, Xu Y, Xu T, Hoy C, Handwerker D, Chen G, Northoff G, Zuo X, Bandettini P (2014). Brain network informed subject community detection in early-onset schizophrenia. Scientific Reports 4, 5549. full text
INCloud and WeProMRITM

- INCloud provides a full-stack solution for the entire process of large-scale neuroimaging data collection, management, analysis, and clinical applications.
- It helps standardize the acquisition and pre- processing of neuroimaging data, making it easier to aggregate data collected by multiple teams.
- INCloud implements a brain feature library that allows users to query, manage, analyze and share brain features, and therefore accelerates the utilisation of neuroimaging data.
- WeProMRITM is our implementation of a user-friendly website for automatical, parallel processing of multimodal brain MRI data. The pro-version of PHI-pipe with more functionalities underlies this website.
- Due to limited compuational resources, please contact us (mricenter#at#smhc.org.cn) for accessing INCloud and WeProMRI website.
References
Li Q, Jiang L, Qiao K, Hu Y, Chen B, Zhang X, et al. (2021): INCloud: integrated neuroimaging cloud for data collection, management, analysis and clinical translations. Gen Psych 34: e100651. full text
